Learning Objectives
- Know that while single gene diseases are rare, they do occur. Explain how a dominant or recessive single gene disease could be inherited, and give some examples of single-gene diseases.
- Define enzyme pathways, explain how gene products can interact in a pathway, and recognize that a single mutation can impact a pathway to cause disease.
- Define polygenic complex trait, recognize polygenic traits by their continuous trait distribution, and read a complex Punnett square to calculate the probability of having offspring for each phenotype.
- Know some examples of disease risk genes, the factors that enhance or decrease risk, and consider the steps you would take to research and learn more about a certain disease if the need arises.
Single gene diseases are rare
Remember Punnett squares? We can use them to explain how a single gene disease could be inherited. For example, sickle cell disease occurs when a person has two mutant copies of the hemoglobin gene. Hemoglobin’s normal function in a red blood cell is to bind oxygen for transport to cells throughout the body. A single base change to the genetic code changes one amino acid in the hemoglobin protein. Because of this single mutation, the altered hemoglobin molecules form long fibers that distort the disc-shaped red blood cells into a crescent shape. The cells with crescent or “sickled” shapes cluster and stick in the smallest blood vessels, called capillaries, preventing effective oxygen transport. The sickled blood cells undergo apoptosis more rapidly than normal blood cells. The low oxygen leads to symptoms of anemia, chronic pain, organ damage, and shortened lifespan.
If a disease condition occurs because of loss of function (where a protein stops working due to a mutation in the gene that encodes it), like sickle cell, then having two copies of each gene buffers us against single mutations causing disease. As long as one copy is normal, the normal protein can still carry out its job. However, if both copies become non-functional, then the full condition or disease will manifest. For this reason, most loss of function conditions are usually recessive: alkaptonuria, phenylketonuria, albinism, cystic fibrosis, and sickle cell anemia are all recessive conditions. In some cases, having only one functional copy reduces function, so heterozygotes for, say, sickle cell are often anemic because they don’t circulate sufficient oxygen, but they are not severely debilitated.
Traits are complex, influenced by gene pathways, polygenic inheritance, and environmental effects
[Modified from Khan Academy]
While single gene diseases do exist, they are rare. Most traits are influenced by multiple genes and by the environment in which those genes are expressed, including many characteristics important in our everyday lives, such as height, skin color, eye color, and risk of diseases like diabetes. We’ll take these ideas in turn.
-
Enzyme pathways. An enzymatic pathway is a series of connected chemical reactions that feed one another. The pathway takes in one or more starting molecules and, through a series of intermediates, converts them into products.
In general, each gene encodes a single enzyme that carries out some function in the cell. Enzymes work together to complete tasks in the cell. For instance, a three enzyme pathway is required to convert a starting substrate (green triangle) into the final product needed by the cell (yellow box).
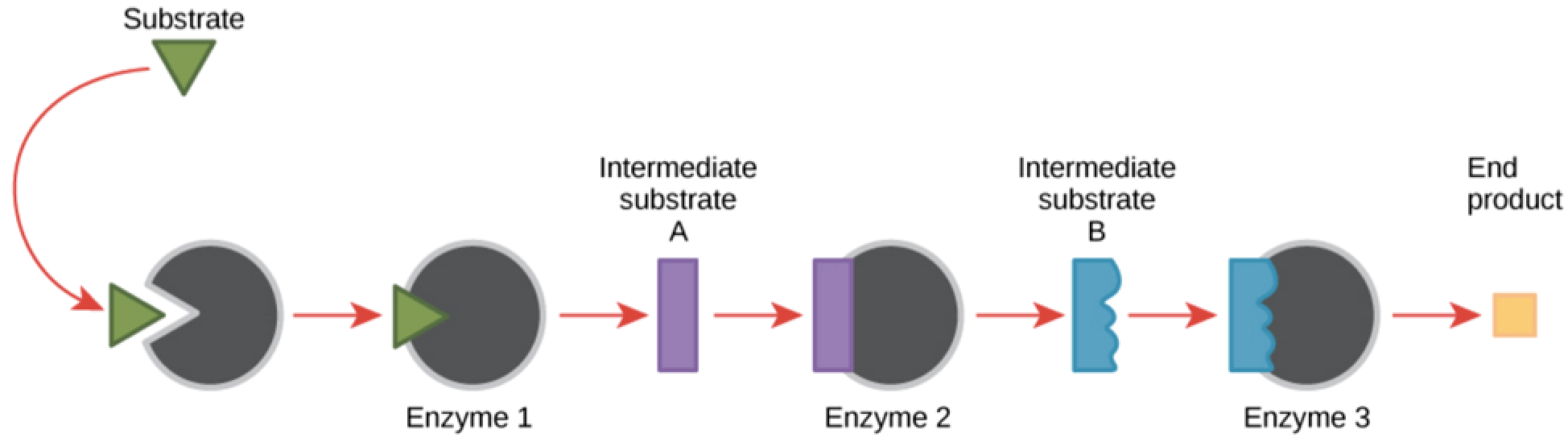
Image credit: OpenStax Biology.
Each enzyme in the pathway converts the substrate into a different intermediate form like an assembly line. If one of those enzymes is missing, the assembly line stops, and the end product needed by the organisms cannot be made. That missing end product could affect the health of the organism, causing disease.
Because each of these enzymes is encoded by a different gene, a single mutation could cause disease by preventing formation of the correct end product. In reality, these pathway traits work pretty much like single gene traits in that if one enzyme’s gene is mutated, the phenotype is affected. Sometimes an intermediate substrate works well enough or is partially functional.
- Polygenic inheritance. Some characteristics are polygenic, meaning that they’re controlled by multiple different genes (not just two). In polygenic inheritance, traits often form a phenotypic spectrum, like heights or pigmentation, rather than falling into clear-cut presence/absence of a trait.
Looking at a real example of a human polygenic trait would get complicated, largely because we’d have to keep track of tens, or even hundreds, of different allele pairs (like the 400 involved in height!). However, we can use an example involving wheat kernels to see how several genes whose alleles “add up” to influence the same trait can produce a spectrum of phenotypes [1,4].
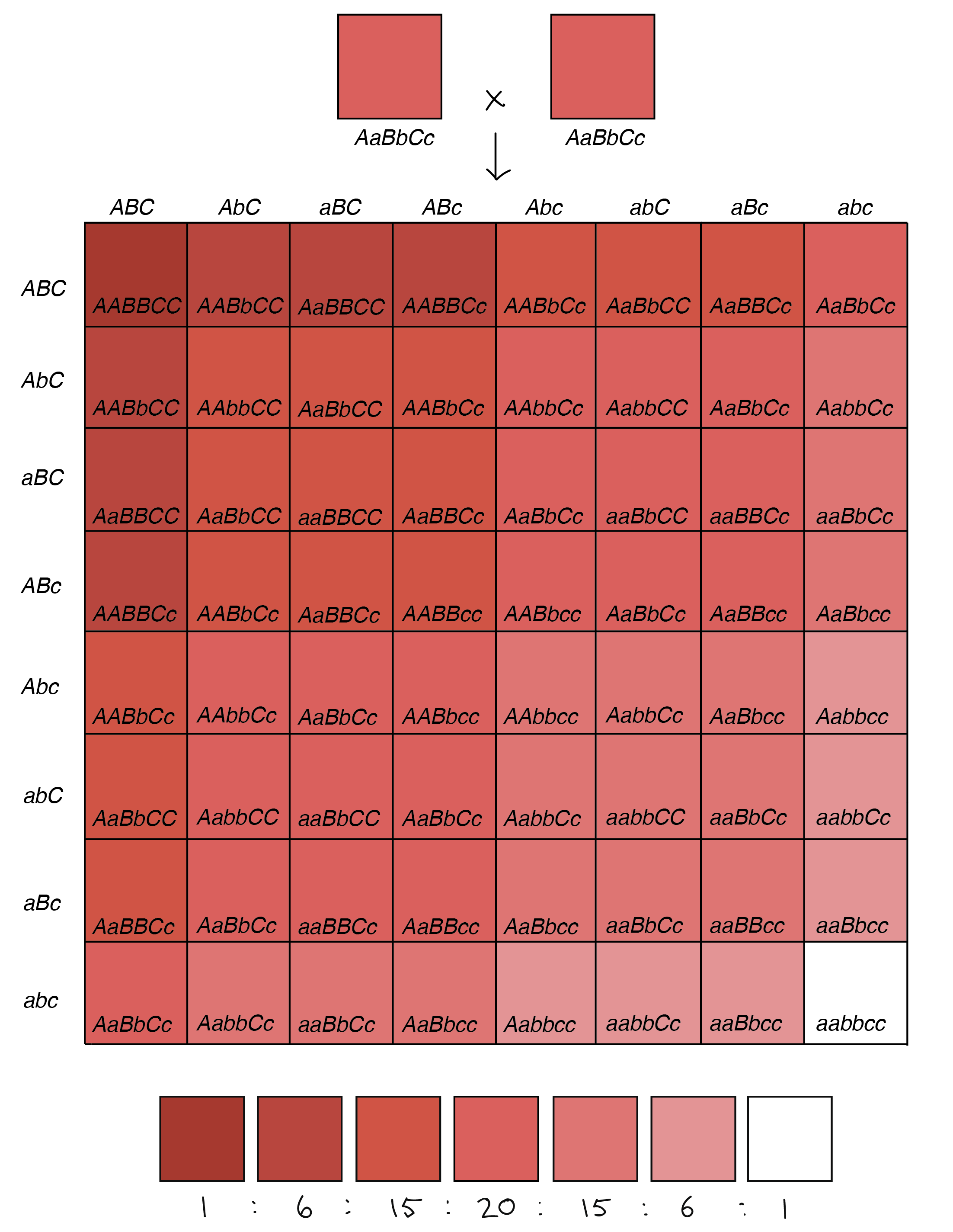
64-square Punnett square illustrating the phenotypes of the offspring of an AaBbCc x AaBbCc cross (in which each uppercase allele contributes one unit of pigment, while each lowercase allele contributes zero units of pigment).
Image credit: Khan Academy. Diagram based on similar diagram by W. P. Armstrong [5].
Now, let’s imagine that two plants heterozygous for all three genes (AaBbCc) were crossed to one another. Each of the parent plants would have three alleles that made pigment, leading to pinkish kernels. Their offspring, however, would fall into seven color groups, ranging from no pigment whatsoever (aabbcc) and white kernels to lots of pigment (AABBCC) and dark red kernels. This is in fact what researchers have seen when crossing certain varieties of wheat [1,4].
This example shows how we can get a spectrum of slightly different phenotypes (something close to continuous variation) with just three genes. It’s not hard to imagine that, as we increased the number of genes involved, we’d be able to get even finer variations in color, or in another trait such as height.
- Environmental effects. Most real-world characteristics are determined not just by genotype, but also by environmental factors that influence how genotype is translated into phenotype.
Reflect back on the sickle cell anemia single-gene disease example. It seems like a simple one-gene disease, but in fact the sickle cell allele conveys different fitnesses in different environments. When the Plasmodium parasites infect and develop in human red blood cells, they cause malaria. But programmed cell death removes the sickled red blood cells from the blood before malaria can develop, so a sickle cell heterozygote tends to have less severe malarial infections in regions where Plasmodium occurs.
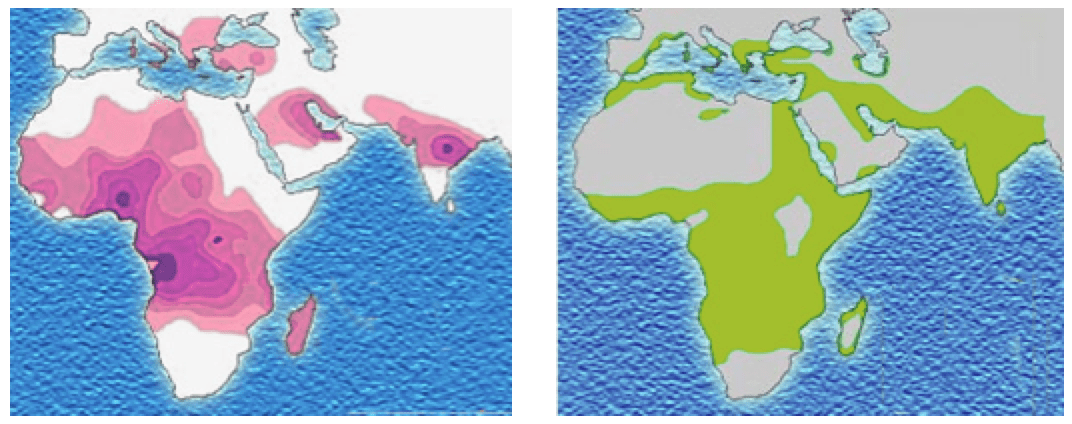
Distribution of the sickle-cell trait shown in pink and purple. Historical distribution of malaria (no longer endemic in Europe) shown in green. (Image credits: Wikipedia CC-LAYOUT; CC-BY-SA-2.5,2.0,1.0)
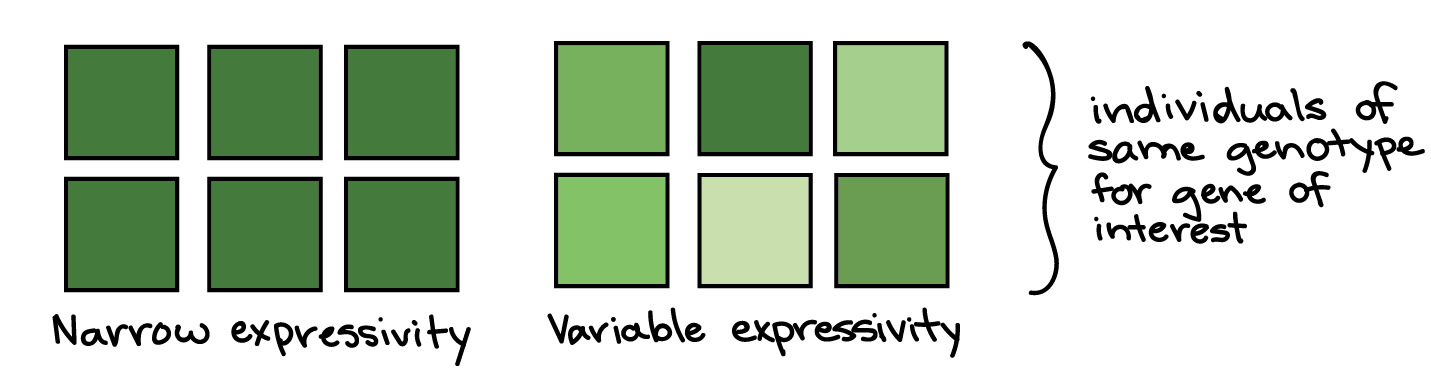
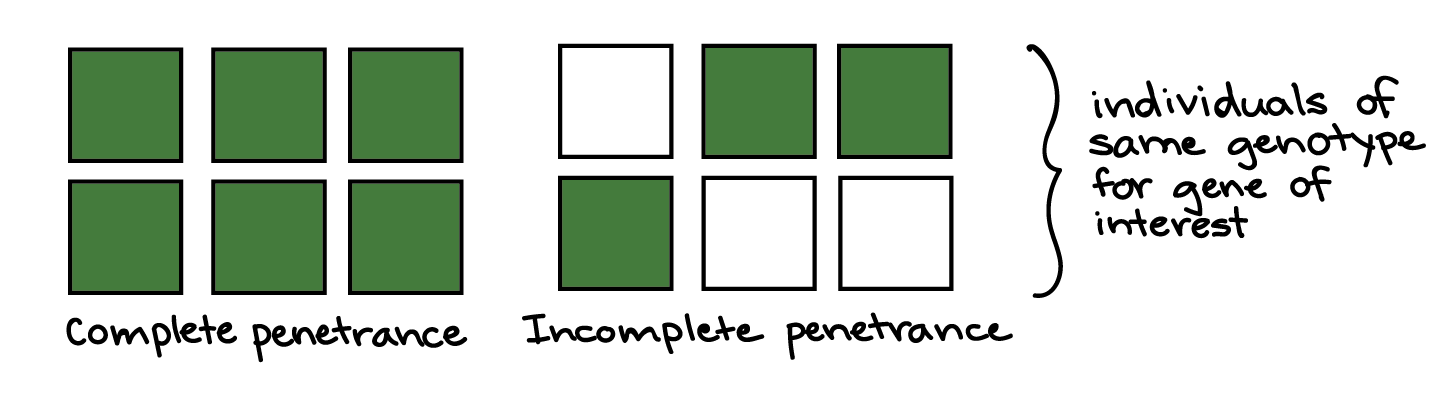
Variable penetrance, incomplete expressivity
[Modified from Khan Academy]
Some genes enhance disease risk or other traits only under certain conditions, called a gene-by-environment interaction. Even for characteristics that are controlled by a single gene, it’s possible for individuals with the same genotype to have different phenotypes. For example, in the case of a genetic disorder, people with the same disease genotype may have stronger or weaker forms of the disorder, and some may never develop the disorder at all.
In variable expressivity, a phenotype may be stronger or weaker in different people with the same genotype. For instance, in a group of people with a disease-causing genotype, some might develop a severe form of the disorder, while others might have a milder form. The idea of expressivity is illustrated in the diagram below, with the shade of green representing the strength of the phenotype.
In incomplete penetrance, individuals with a certain genotype may or may not develop a phenotype associated with the genotype. For example, among people with the same disease-causing genotype for a hereditary disorder, some might never actually develop the disorder. The idea of penetrance is illustrated in the diagram below, with green or white color representing the presence or absence of a phenotype.