Learning Objectives
- Know and use the vocabulary needed to discuss genetic inheritance including gene, allele, dominant, recessive, gamete, genotype, phenotype, homozygote, heterozygote, carrier
- Construct and use a Punnett square for a single trait and for two traits, using appropriate terminology, and use it to determine possible offspring types and phenotypic ratios.
- Define the chromosome theory of inheritance as “genes are located on chromosomes”
- Explain how crossing over and recombination generate new combinations of alleles and new variation in a sexually reproducing population.
The terminology of Mendelian inheritance
Gregor Mendel discovered “particulate inheritance” or the idea that hereditary elements are passed on in discrete units rather than “blended” together at each new generation. Today we call those discrete units genes.
- A gene is a hereditary factor that determines (or influences) a particular trait. A gene is comprised of a specific DNA sequence and is located on a specific region of a specific chromosome. Because of its specific location, a gene can also be called a genetic locus.
- An allele is a particular variant of a gene, in the same way that chocolate and vanilla are particular variants of ice cream.
- An organism’s genotype is the particular collection of alleles found in its DNA. An organism with two of the same alleles for a particular gene is homozygous at that locus; an organism with two different alleles for a particular gene is heterozygous at that locus.
- An organism’s phenotype is its observable traits. An organism can have a heterozygous at a particular locus but have a phenotype that looks like only one of the two alleles. This is because some alleles mask the appearance of others in a dominant/recessive pattern.
- A dominant allele produces its phenotype whether the organism is homozygous or heterozygous at that locus. For example, in humans the allele for brown eyes is dominant to the allele for blue eyes, so a person who is heterozygous at the eye color locus will have brown eyes.
- A recessive allele produces its phenotype only when homozygous at the locus; its phenotype is masked if the locus is heterozygous. For example, a person must have two copies of the blue eye color allele to have blue eyes.
- Sometimes specific recessive alleles are associated with diseases. A person who is heterozygous for the gene will be phenotypically normal, but carry a copy of the recessive, disease-associated allele. This person is said to be a carrier and can pass on the disease allele to his or her offspring.
Crosses with a single trait & the law of segregation
All of the concepts above are illustrated in the types of experiments that Mendel carried out with pea plants. Pea plants aren’t a particularly exciting organism to study, but they were very useful in figuring out basic patterns of inheritance because they have a lot of traits that are caused by a single gene with a simple dominant/recessive inheritance pattern. While single-gene inheritance of major traits is actually pretty rare in general, we’ll learn some good disease examples later in this course, and also some exceptions. But first, let’s get the basics of gene inheritance by learning about single genes and their traits in peas. A classic example is pea shape. Peas can be either round or wrinkly, but not anything in between. Whether they are round or wrinkly is controlled by a single gene with two variants, or alleles: the round allele is dominant to the wrinkly allele. The inheritance pattern if you cross homozygous round and homozygous wrinkly pea plants is illustrated here:

You can see in the first generation (F1) that all offspring produce round seeds, even though they carry both the round and wrinkly alleles. If the F1 generation self-fertilizes (pea plants – like most plants – produce both male and female gametes), then you now see some offspring that produce rounds seeds and some that produce wrinkly seeds. The round:wrinkly seed producers exist in approximately a 3:1 ratio, illustrated by constructing a Punnett square.
Punnett squares illustrate the fact that each pea plant gamete contains only one allele for each trait. Even though adult pea plants have two copies of each allele, these two alleles are separated into different gametes. Thus when any two gametes come together to create a new plant, each gamete carries one allele. The games unite, resulting in two alleles in the new plant. The idea that each gamete carries only one allele for each trait is the law of segregation; that is, the two alleles for a particular trait are segregated into different gametes.
Crosses with two traits and the law of independent assortment
Pea plants have a lot of other traits beyond seed shape, and Mendel studied seven other traits. Things become more complex when you follow more than one trait at at time. Here is a cross looking at both pea shape (round or wrinkly) and pea color (yellow or green). Follow the logic of the cross below to see why offspring demonstrate a 9:3:3:1 ratio of different phenotypes.
Punnett squares that show two or more traits illustrate the idea that alleles for different traits segregate independently of each other. Yellow seeds are not always round, and green seeds are not always wrinkly; there can be yellow wrinkly seeds, yellow round seeds, green wrinkly seeds, and green round seeds. The idea that alleles for different traits segregate independently is the law of independent assortment.
Mendel’s laws and meiosis
Mendel’s laws of segregation and independent assortment are both explained by the physical behavior of chromosomes during meiosis.
Segregation occurs because each gamete inherits only one copy of each chromosome. Each chromosome has only one copy, or allele, of each gene; therefore each gamete only gets one allele. Segregation occurs when the homologous chromosomes separate during meiotic anaphase I. This principle is illustrated here:
Source: Adapted from Biology Dictionary (https://biologydictionary.net/law-of-segregation-vs-law-of-independent-assortment/)Independent assortment occurs because homologous chromosomes are randomly segregated into different gametes; ie, one gamete does not only get all maternal chromosomes while the other gets all paternal chromosomes. Independent assortment occurs when homologous chromosomes align randomly at the metaphase plate during meiotic metaphase I. This principle is illustrated above, where the patterns on the left and right show two independent ways that the little r allele from the round gene can be matched with an allele (y or Y) from the yellow gene:
Random, independent assortment during metaphase I can be demonstrated by considering a cell with a set of two chromosomes (n = 2). In this case, there are two possible arrangements at the equatorial plane in metaphase I. The total possible number of different gametes is 2 to the power of n, where n equals the number of chromosomes in a set. In this example, there are four possible genetic combinations for the gametes. With n = 23 in human cells, there are over 8 million possible combinations of paternal and maternal genotypes in a potential offspring.
Here’s a quick summary of many of these ideas from Ted Ed:
and here is Khan Academy’s take:
Genes are located on chromosomes
Genes are located on chromosomes, which are long linear strands of DNA. Because genes are like beads on the string of DNA, some genes are near to each other, while others are far apart, even on different chromosomes from each other. Those genes on the same chromosome can be so close together that they are physically linked, violating Mendel’s law of independent assortment. Genetic linkage occurs when the genes controlling two different traits are located near each other on the same chromosome. The basic idea is that if two genes are on the same chromosome, and you inherit the whole chromosome, then you have to inherit those two allelic variants of those genes together.
However, this is biology so there is a caveat: the phenomenon of crossing over helps to shuffle the alleles for genes located on the same chromosome. A crossover event between the locations of two genes on a chromosome results in genetic recombination, or new combinations of alleles on a chromosome.

Crossing over between genes A and B results in recombinant chromosomes with new allele combinations a, b and A, B, in addition to the original parental combinations A, b and a, B. Image from Wikimedia by user Abbyprovenzano, with CC-BY-SA-3.0 license.
Crossing over occurs during meiotic prophase I, when the homologous chromosomes align and synapse, and results in physically swapping genetic material (DNA) between non-sister chromatids of the paired homologous chromosomes. Because crossing over occurs randomly along the chromosome, the closer two genes are physically located to each other on a chromosome, the less likely that a crossover will occur between them. Conversely, the farther apart two genes are located from each other along the chromosome, the more likely they are to be swapped with the alleles on the homologous chromosome. The image below illustrates this idea:
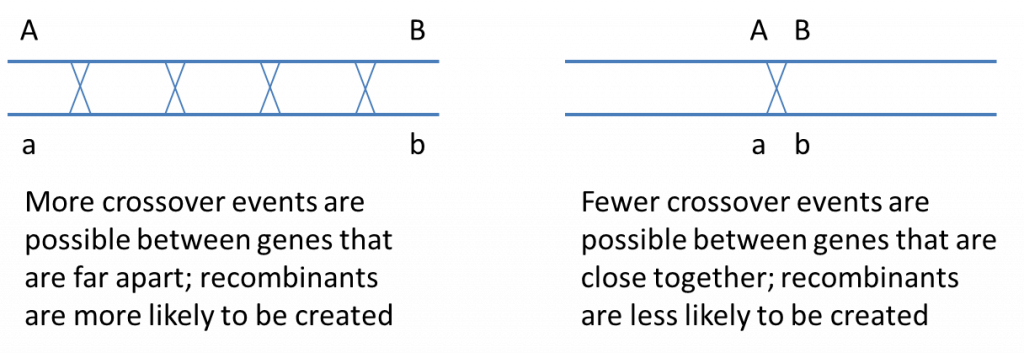 It may be surprising to realize that two genes on the same chromosome will assort independently (like genes located on separate chromosomes) if they are far enough apart that a crossover almost always occurs between them, producing 50% recombinants (because crossing over involves only two of the 4 chromatids in a synapsed pair of homologous chromosomes, the maximum recombination frequency is 50%).
It may be surprising to realize that two genes on the same chromosome will assort independently (like genes located on separate chromosomes) if they are far enough apart that a crossover almost always occurs between them, producing 50% recombinants (because crossing over involves only two of the 4 chromatids in a synapsed pair of homologous chromosomes, the maximum recombination frequency is 50%).
The video below walks through linkage as a violation of independent assortment and explains how crossing over breaks linkage. Note this video uses an incomplete definition of linkage: linkage occurs when two genes are located close together on the same chromosome and thus tend to be inherited together. It is not sufficient for genes to be on the same chromosome to be linked; they also have to be close enough together that crossing over between them is a relatively rare event.